Little Giants
A closer look at a tiny bug with a big role to play
Michael Pfrender sits facing a whiteboard in his lab at the Galvin Life Science Center. He’s discussing the genomics of Daphnia – water fleas, found in every standing body of water in the world – and has a tendency to sketch when he speaks.
“You want to see some of them?” he asks. “That’s the fun part, right?”
At the back of the lab, two beakers sit on a table near a microscope. There isn’t much to see at first glance. Even leaning in, Daphnia are so small they look like bouncing flecks in the water, frantically trying to keep afloat.
Pfrender dips an eyedropper into one of the beakers, releases a drop of water onto a slide and slips it under the microscope for a closer look.
There she is.
Portly with a round, neckless head and antennae sprouting from either side, she is translucent – her organs visible within the razor-thin outline of her body. She also happens to be pregnant, her eggs equally visible, and encased in little pouches.
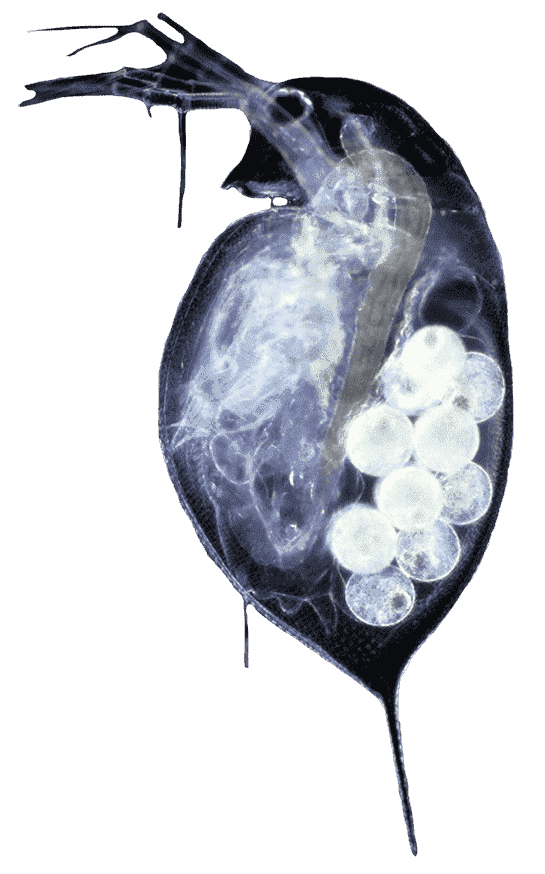
Pfrender, associate professor of evolutionary and ecological genomics in the Department of Biological Sciences and the director of the Genomics and Bioinformatics Core Facility, has studied the genomics of Daphnia for more than two decades. He co-authored the landmark paper documenting the first genome sequence of a Daphnia species – as well as the second, new and improved genome sequence of Daphnia pulex (D. pulex) published earlier this year.
These tiny water fleas play a big ecological role in lakes, ponds and standing waters all over the world. Their sensitivity to chemical cues and ability to adapt to various environments have fascinated scientists for more than 150 years.
Here are some things to know about Daphnia…
They have a long history of scientific study, dating back to the 1600s, when they were known as “the water blob.” It was an exciting discovery. Before microscopes, freshwater biologists didn’t know the organisms even existed in the very lakes they had been studying.
Once they got a glimpse, they were hooked.
“People spent a lot of time describing their anatomy and trying to figure out what they were doing,” Pfrender said. “They started to keep them in a lab and ask questions.”
“Whatever environment they’re in, they seem to adapt and persist.”
Daphnia feed on algae and phytoplankton, playing a vital role in preventing overgrowth – such as the massive algae blooms in 2014 that contaminated drinking water in Toledo, Ohio – that can turn water toxic.
They‘re female – and they’re clones. In ideal conditions, Daphnia reproduce clonally and always produce daughters. The process takes just 12 days. Under strained conditions, with limited food sources or an extreme change in temperature, they make what Pfrender calls “functional males” – identical to the mother to continue reproduction.
They are the primary grazers in any standing body of water and the primary forage for the fish that are next in line in the food chain. Without them, aside from the potential for algae overgrowth, there would be no food for smaller fish.
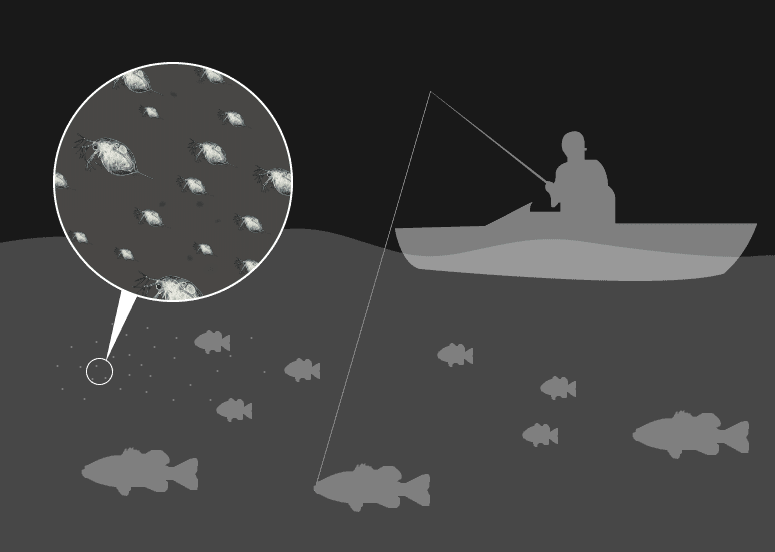
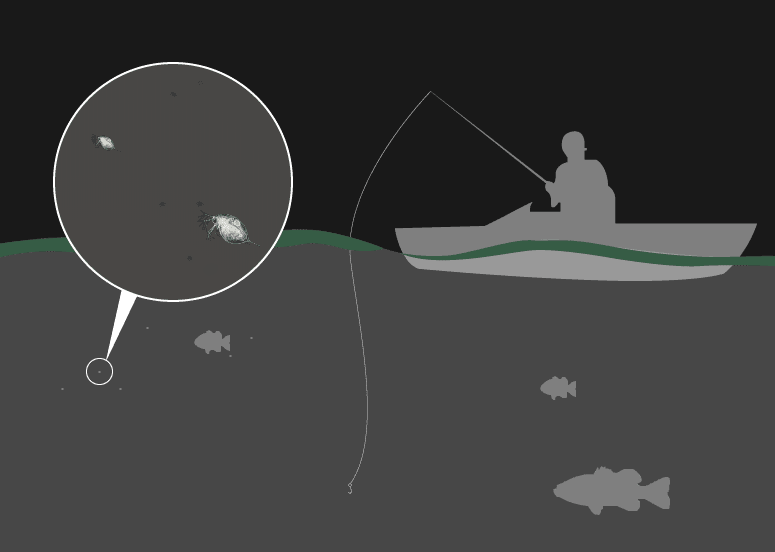
“They sit right in the middle of the trophic food web in lakes, and they are a primary driver of the ecological dynamics of these systems,” Pfrender says.
They are the canaries of the water – so sensitive to their environment the EPA uses them as indicators of acute toxicity in freshwater.
“Whatever environment they’re in, they seem to adapt and persist,” Pfrender says.
Scientists first noted the Daphnia’s unique phenotypic plasticity – modifying the body to the environment – in the early 20th century around 1907.
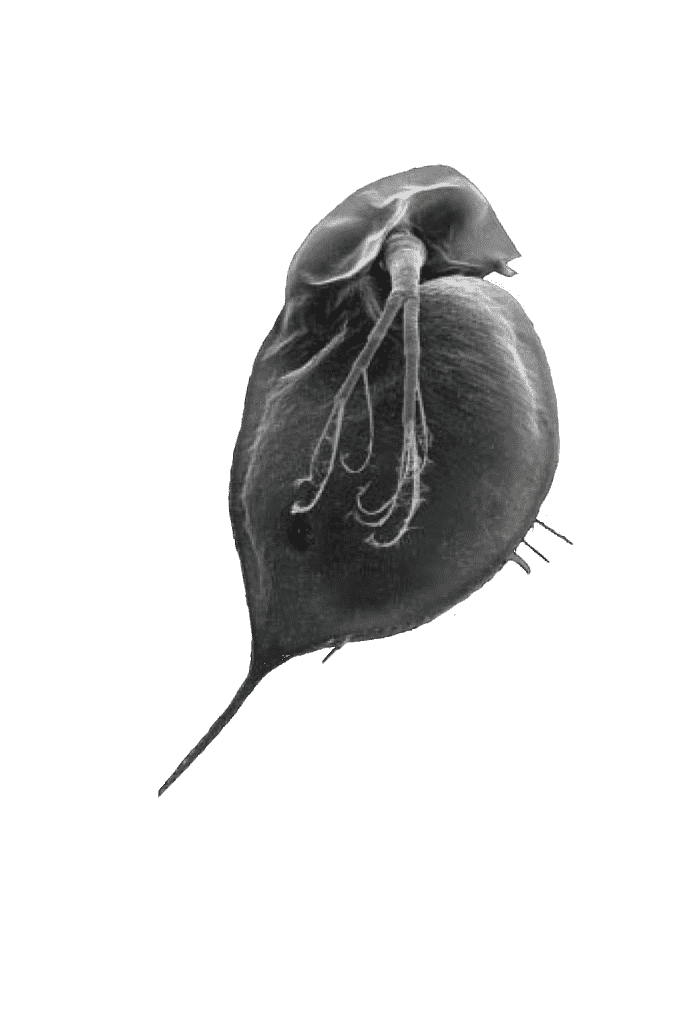
In environments where no predators are present, Daphnia look like those in the beakers in Pfrender’s lab. Rounded, portly, bobbing up and down in the water.
In predatory environments, they develop defensive structures – pointed helmets and elongated spears at the base – making them harder to eat. Some are smaller, lighter in color and less visible to predators.
Pointed Helmet
Elongated Spear
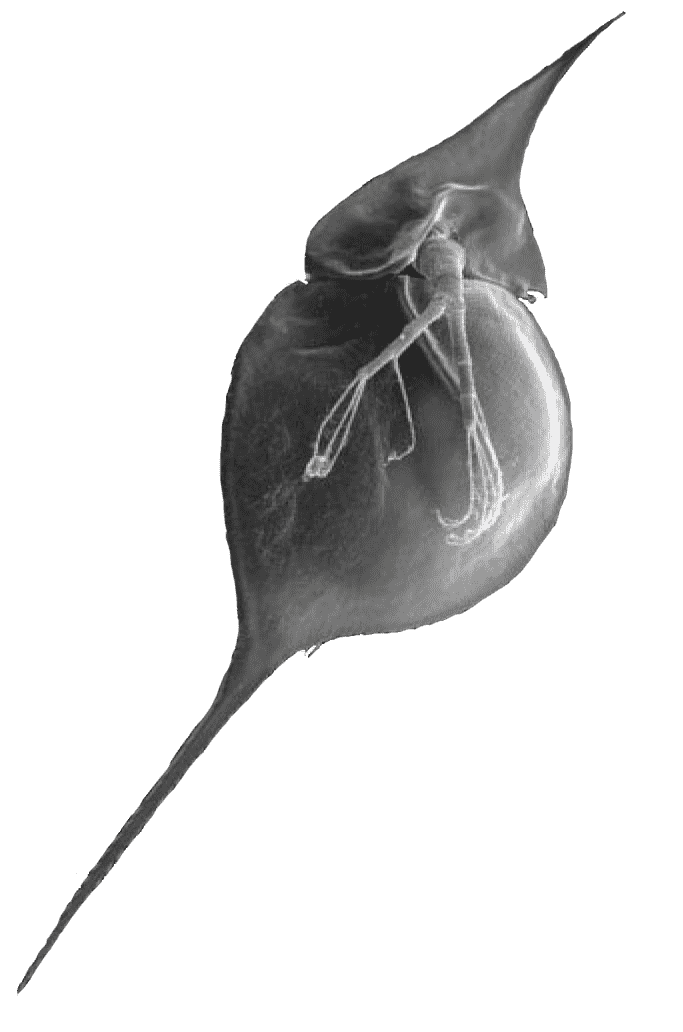
They know when to move. Fish know their prey have to be near algae in order to graze and zero in on those areas during the day. Daphnia know this too, and will move to the bottom of lakes or ponds during the day, rising at night to avoid predators while they feed – a behavior known as diel vertical migration.
These tiny water fleas have a remarkable capacity to live in unique and vastly different environments, like the high saline rock ponds of Churchill Bay in Manitoba. Populations living in the desert ponds of Australia develop a resistance to exposure to high amounts of ultraviolet light. Unlike their translucent relatives, they are black in color.
There are species living in toxic waters full of cyanobacteria and even in ice ponds of the Arctic.
Despite their unique ability to adapt, some species have been forced out by environmental change – which strikes at the heart of Pfrender’s quest to understand the genomic infrastructure of these tiny, not-so-great swimmers, which are really rather cute once you get to know them.
How do some species survive in certain environments while others don’t?
The genome is the operations manual for every living organism.
Researchers began looking at the genetic makeup of Daphnia for the first time when a variety of genetic techniques became available in the 1970s and ’80s. It was a labor intensive and expensive process.
In the late ’90s, fueled by a desire to decode the human genome, genome centers became the only way to generate the massive data of a complete sequence. Through these centers, scientists could sequence whole genomes of model species, comparing genetic variation in populations to look for which functions triggered specific genes.
Pfrender was one of five scientists hoping to sequence a Daphnia genome. The group included researchers from Indiana University, the University of Oregon, the University of New Hampshire and the University of California Davis. It was 2003.

They turned to the Joint Genome Institute (JGI).
“The Joint Genome Institute was one of the original genome centers for the human genome, and after the human genome was completed they were looking for their niche,” Pfrender said. “They really took up the vanguard of genomics of vital organisms that nobody else would do.”
There was a focus on evolutionary development and diversification of species. Researchers were sequencing agriculturally important insects and vectors pertinent to human health and disease, like the honeybee, the fruit fly and the mosquito.
“What was completely missing was an ideal ecological model for environmental science,” Pfrender said. “We thought that was our niche.”
The group pitched the importance of Daphnia to its ecosystems and pointed to a wealth of ecological literature on the water flea – perhaps larger and more detailed than any other aquatic organism.
The pitch was solid. After a short pause the director of the institute asked, “When do we get started?”
The first Daphnia genome sequence was complete in 2006. The team chose a population found in Oregon, favored for its low variation rate. They called it The Chosen One (TCO). The results were published in Science in 2011.
Pfrender had no plans to stop at one. Six years later, he published a new and improved sequence for the D. pulex, a population found in the Midwest, and provided a high-density genetic map in high resolution and greater detail.
These tiny water fleas play a big ecological role in lakes, ponds and standing waters all over the world.
“When you sequence a genome, what you have is a lot of short pieces of sequence that’s like a big jigsaw puzzle and you’re trying to put them together,” says Pfrender. “You want the biggest stretch of DNA that you can get out of these little pieces. How long they are, how evenly spaced – it all comes into play. With this [second] sequence, the Genomics and Bioinformatics Core here at Notre Dame was able to employ new techniques including long-synthetic sequences, which allows us to bridge longer pieces together.”
“The Genomics Core made a lot of the data that went into this assembly,” Pfrender says. “Now we have much longer pieces of continuous information for this genome than we did for the last one. We were also able to connect them to chromosomes so that we know where the jigsaw puzzle pieces sit on each chromosome. This gives us a road map of the structure of the genome.”
Back in the lab, Pfrender dips the eyedropper into another beaker.
These Daphnia had been living in water where a certain bacteria had started to grow, he explains. In response, they began producing hemoglobin. He slides one of these water fleas under the microscope.
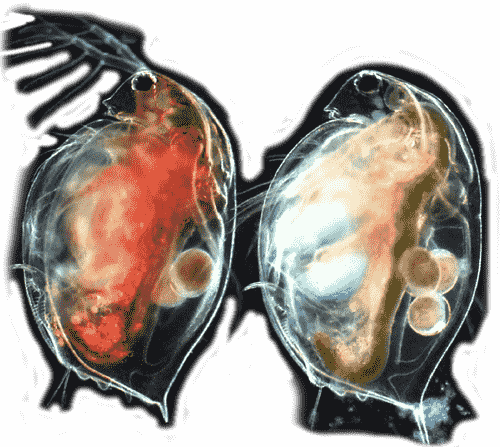
Though she’s genetically identical to the others, and same in shape and size, the hemoglobin coursing through her gives her body a bright red tint.
Pfrender hopes to put his latest genetic map of these water fleas to good use, to identify how Daphnia adapt to their environments and understand these processes as a foundation for human health.
“There are more than 100,000 manufactured chemicals used in the environment. We know the toxicity of fewer than 5 percent, and we know virtually nothing about the effects of exposure to complex mixtures of these compounds,” Pfrender says. “Our own exposure to those chemicals may have very subtle effects that only show up years later. If we can reveal the genomic underpinnings, and understand the responses of an organism like Daphnia to multiple stressors, we can move toward a predictive science informed by data that could affect human health around the world. This is a great challenge for the 21st century.”